- Home
- About Us
- Service
- Application
- Technology
- Resource
- Contact Us

Whole Transcriptome Sequencing
Whole Transcriptome Sequencing (RNA-Seq) is a sophisticated technique that sequences all RNA transcripts in a sample, providing detailed insights into gene expression, novel transcripts, and alternative splicing. Widely used in fields like oncology and neurology, RNA-Seq helps researchers understand complex biological processes and identify disease biomarkers with high sensitivity and resolution.
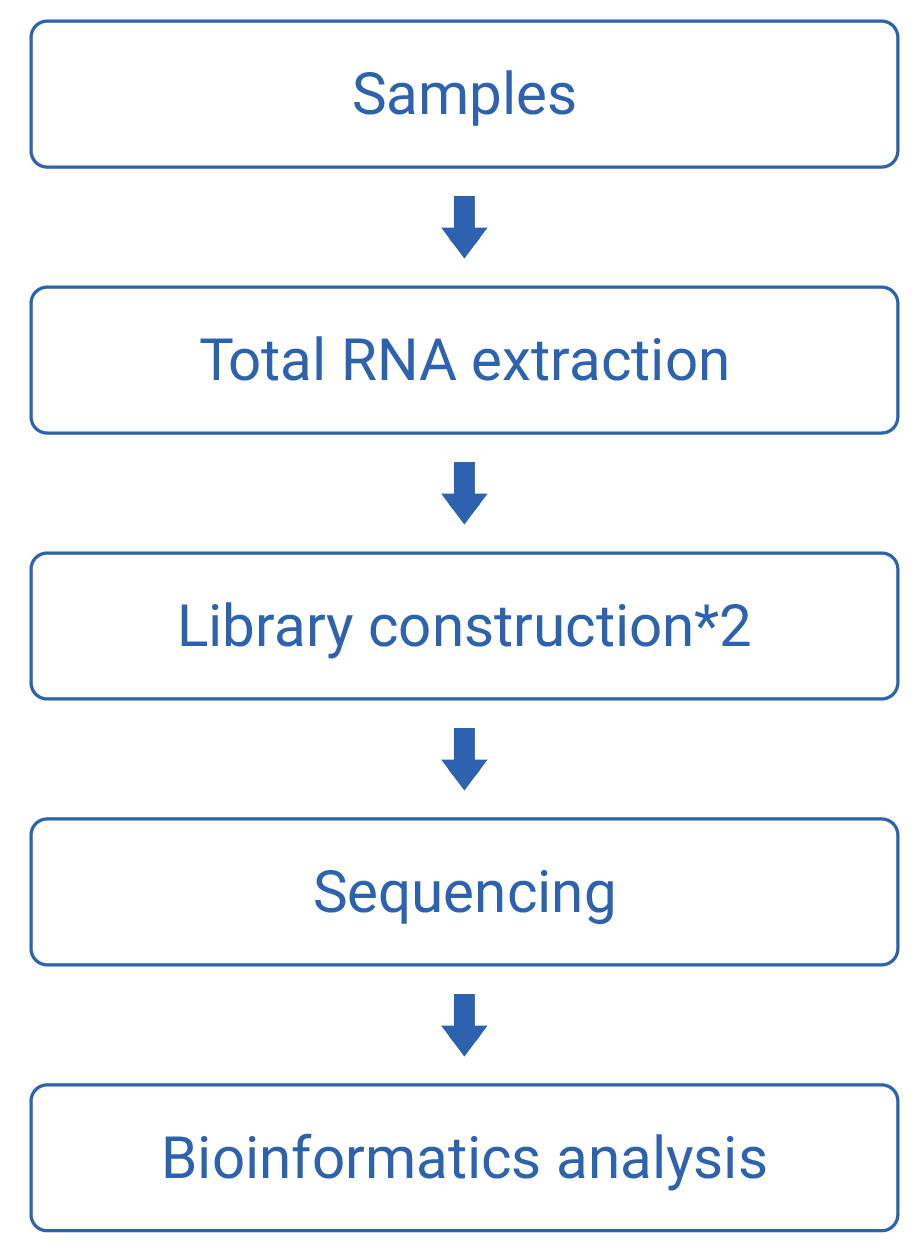
Work flow
Technical Parameters
| Sequencing range | lncRNA+circRNA+mRNA+miRNA |
| Sequencing strategy | NGS PE150 |
| Sequencing throughput | 15Gb lncRNA+circRNA+mRNA3Gb miRNA |
| Data quality | Fastq files, Q30≥85% |
| Data analysis | Standard analysis |
| TAT | Standard: 35 WD |
Applications
| Gene Expression Profiling | Alternative Splicing and Isoform Discovery | Transcriptome Annotation |
| Disease Research and Biomarker Discovery | Non-coding RNA Characterization | Developmental Biology |
Bioinformatics Analysis
1.Data Quality Control: The quality of sequencing data is a crucial prerequisite for the reliability of information analysis results. It is necessary to undergo a rigorous quality control process on post-sequencing data to obtain high-quality sequencing data.
2.Basic Whole Transcriptome Analysis: Perform basic analysis on mRNA, lncRNA, circRNA, and microRNA respectively, to identify subsets of differentially expressed molecules.
3.Whole Transcriptome Association Analysis: Based on miRNA target molecule prediction and MRE identification, perform whole transcriptome regulatory network analysis.
- 3.1 Prediction of miRNA target molecules
- 3.2 Statistics on miRNA Response Elements
- 3.3 miRNA-lncRNA Association Analysis
- 3.4 miRNA-circRNA Association Analysis
- 3.5 miRNA-mRNA Association Analysis
- 3.6 lncRNA-mRNA Association Analysis
- 3.7 circRNA-mRNA Association Analysis
- 3.8 lncRNA-miRNA-mRNA Association Analysis
- 3.9 circRNA-miRNA-mRNA Association Analysis
- 3.10 lncRNA-circRNA-mRNA-miRNA Association Analysis
Advantages
![]() High Sensitivity
High Sensitivity
![]() Comprehensive Coverage
Comprehensive Coverage
![]() Bias-Free Analysis
Bias-Free Analysis
