- Home
- About Us
- Service
- Application
- Technology
- Resource
- Contact Us

Meta 16S/18S/ITS Sequencing
16S/18S/ITS ribosomal RNA (rRNA) sequencing is a specialized high-throughput sequencing technology designed to analyze the 16S/18S/ITS rRNA gene, a component of the 30S small subunit of prokaryotic ribosomes found exclusively in bacteria and archaea. This gene is highly conserved across different species of bacteria and archaea, making it an ideal target for microbial identification and phylogenetic studies.
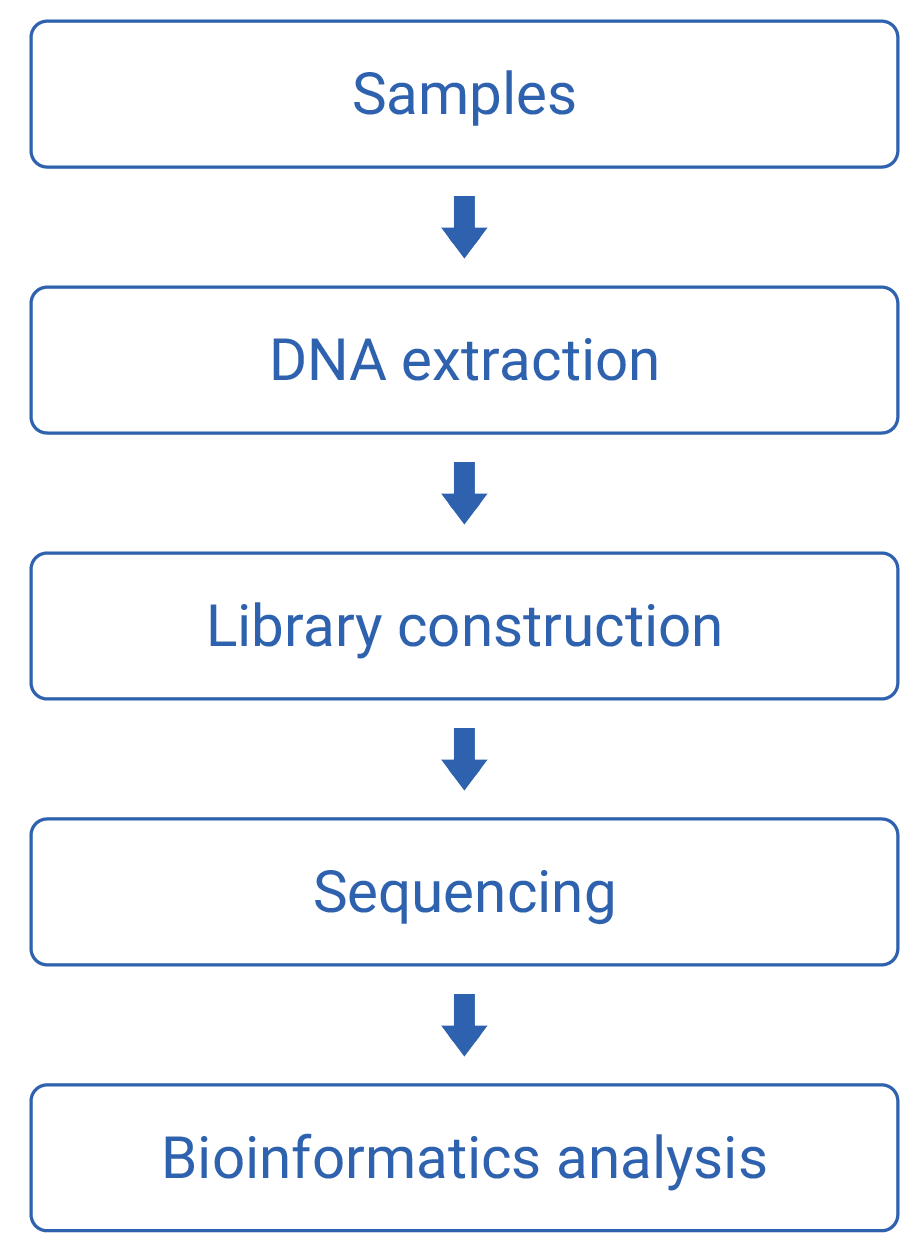
Work flow
Technical Parameters
| Sequencing range | Specific regions (16S/18S/ITSregions) |
| Sequencing strategy | NGS PE250 |
| Sequencing throughput | 30K reads |
| Data quality | Fastq files, Q30≥80% |
| Data analysis | Standard+advanced analysis |
| TAT | Standard: 25 WD |
Applications
| Environmental Microbiology | Agriculture and Soil Science | Antibiotic Resistance Monitoring |
| Human Microbiome Research | Food Safety and Fermentation Industry | Bioremediation and Ecological Restoration |
Bioinformatics Analysis
1.Data Quality Control: Remove primer sequences, filter low-quality sequences.
2.OTU Clustering: OTU sequences, OTU abundance table, inter-group OTU comparison (Venn diagram).
3.Species Annotation: OTU species annotation, interactive heatmap of annotation results, multi-level species composition diagram, species composition bar chart, species abundance clustering heatmap, genus-level phylogenetic tree.
4.Alpha Diversity Analysis: Diversity index statistics, diversity index differential analysis, rarefaction curve, Rank-Abundance curve.
5.Beta Diversity Analysis: Diversity index differential analysis, PCA analysis, PCoA analysis, NMDS analysis, UPGMA clustering tree.
6.Significance Testing of Community Structure Differences Between Groups: ANOSIM analysis, MRPP analysis, ADONIS analysis, AMOVA analysis.
7.Differential Analysis: Metastats analysis, Lefse analysis.
8.Functional Analysis: Relative abundance of functional annotations, clustering analysis of functional relative abundance, PCA analysis of functional annotations, Circos diagram of functional annotations, KEGG differential analysis.
9.Correlation Analysis: Network analysis, dynamic network diagram.
Advantages
![]() High Specificity
High Specificity
![]() Broad Applicability
Broad Applicability
![]() Standardized Procedures
Standardized Procedures
